Highlight
New Tracking Algorithm for the Study of Cell Motility
Achievement/Results
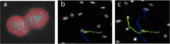
A team composed by Prof. James Henderson (Bioengineering) and Prof. Lisa Manning (Physics), bioengineering graduate students Megan Brasch and Richard Baker and IGERT trainee Forrest Smith (physics) has developed a new automated approach that accurately and efficiently tracks cells in complex in vitro environments over long time scales (tens of hours or days). The code will enable quantitative analysis of cell motility in dense collections of cells moving in diverse complex environments, such as substrates of controlled topology. The method is based on a new tracking algorithm that allows the identification of stained cell nuclei in low contrast images. Most importantly, it includes a technique for recognizing cell interactions, such as divisions and merging events.
This is an important new element for studying dense cell colonies, as many existing automated particle and cell tracking software are unable to distinguish closely interacting cells and therefore face problems when analyzing biological cells at intermediate or high densities. The new “fingerprint” cell tracking software developed by the team overcomes these limitations by using a contour-based approach. This approach identifies variably labeled cells and requires no intensity threshold, quickly pinpoints cell division events, and tracks individual cells through these events using shape identifiers. The algorithm was first tested on synthetic data of cells moving on a grooved substrate obtained via a previously developed code for simulating the dynamics of dense collections of cells in two dimensions. It has also already been used to track living cells and measure mean-square displacements.
Address Goals
The new tracking algorithm will allow us to characterize quantitatively the dynamics of dense cell colonies and to identify novel length and timescales that are important for collective cell behavior. Recent research into cell-substrate interactions has examined how single cell motility and protein expression is influenced by nanopatterned substrates, substrates with different stiffnesses, and substrates with different chemical composition. These studies have all, however considered the response of cells to variations in the environment that are stationary in time. In contrast, many inter- and intra-cellular processes occur over a very wide range of timescales: cytoskeletal rearrangements can occur over less than a minute, while changes to gene expression often occurs over hours or days. This suggests that there is an opportunity to tease apart which of these microscopic properties are influencing collective cell motility by changing the substrate properties over time in a controlled, pre-programmed way. In particular, we hypothesize that collective cell motility will be quantitatively different depending on whether an underlying substrate changes topography quickly (over seconds to minutes) or slowly (over hours to days). Our tracking software will allow us to verify this hypothesis.
The project will provide a freely available, transferable software package that accurately tracks variably-stained cells or nuclei through close interactions and divisions. While the contour algorithm is more computationally intensive than existing routines, it fills an important gap in the tracking literature for situations where information is needed over long timescales at high cell densities.